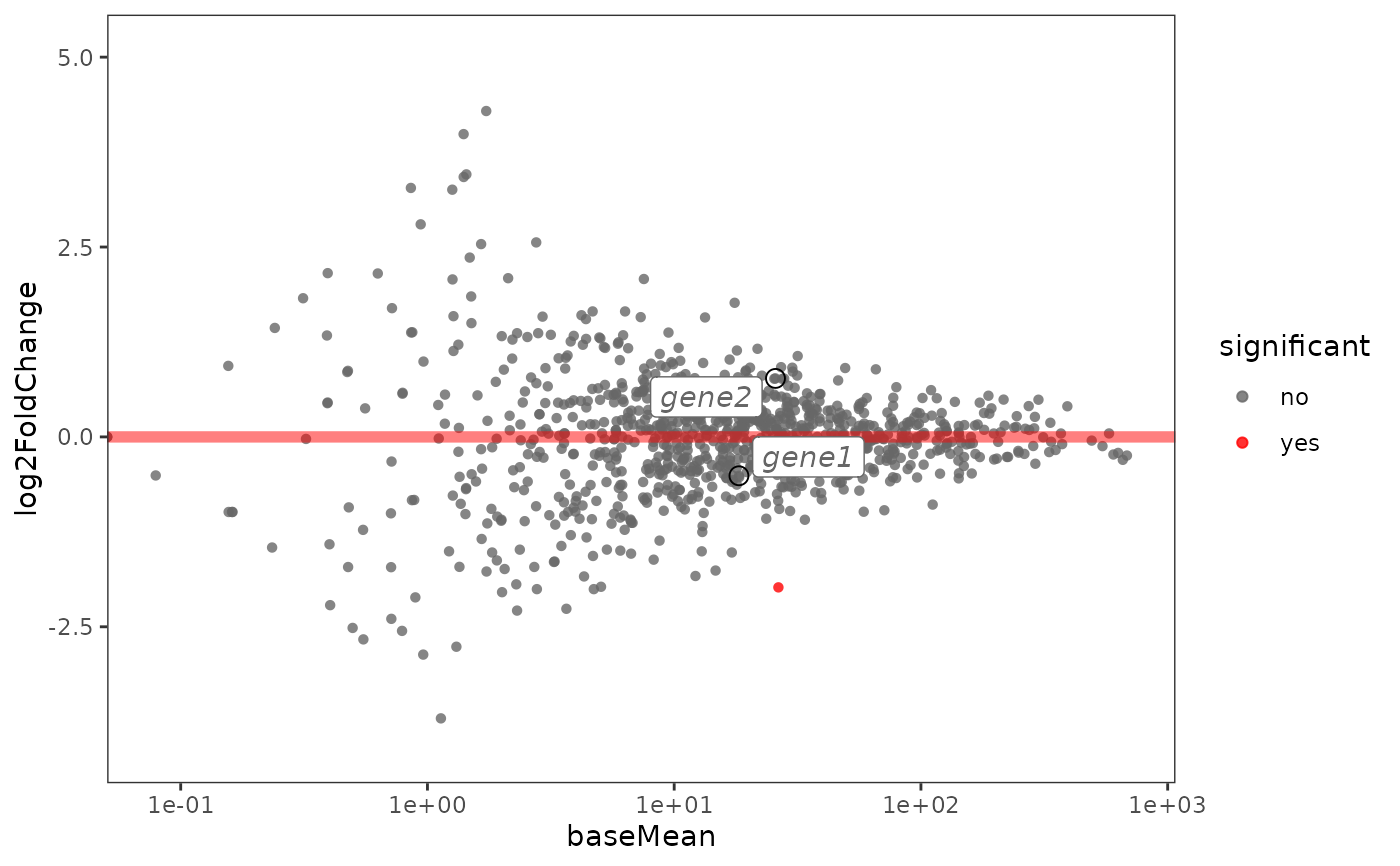
This function creates an MA plot from a data.frame containing DE analysis results.
Usage
plotMA.label(
res,
fdr.thres = 0.01,
fc.thres = 0,
fc.lim = NULL,
lab.genes = NULL,
tolower.cols = c("SYMBOL", "ALIAS")
)Examples
library(DESeq2)
# make example DESeq dataset
dds <- makeExampleDESeqDataSet()
# run DE analysis
dds <- DESeq(dds)
#> estimating size factors
#> estimating dispersions
#> gene-wise dispersion estimates
#> mean-dispersion relationship
#> final dispersion estimates
#> fitting model and testing
# extract comparison of interest
res <- results(dds, contrast = c("condition", "A", "B"))
# add gene and symbol column
res$gene <- rownames(res)
res$symbol <- rownames(res)
plotMA.label(res, lab.genes = c("gene1", "gene2"))
#> Warning: log-10 transformation introduced infinite values.